Epigenetics in MS: A Primer
Epigenetic changes may account for much of the heterogeneity in MS, both in disease onset and progression
The most vexing and enduring question in multiple sclerosis research may be: Why do some people develop MS when others do not? Over the years, researchers have identified a number of genetic and environmental triggers, but an individual may possess every one of those risk factors and never develop the disease. Another patient may have only one or two of the risk factors and still have MS.
A growing body of literature suggests that epigenetic modifications—heritable changes in gene expression that do not involve a mutation in the DNA itself—may hold the keys to understanding why some develop MS while others do not.
Proponents of this idea argue the case on multiple levels. MS is passed down more often by mothers than fathers, and that could potentially be explained by an epigenetic change on the leukocyte antigen-DRB1*15 allele (Küçükali et al., 2014). MS also has a concordance rate of between 30% and 40% in monozygotic twins and only 3% to 5% in dizygotic twins; epigenetic modifications may also explain this difference (Koch et al., 2013). Additionally, the three major environmental risk factors—Epstein-Barr virus (EBV) (see “Viral Villain”), vitamin D deficiency (see “The Sunshine Suspect”), and smoking (see “Where There’s Smoke (and Stress), Is There Fire?”)—are all known to result in pathogenic changes to the epigenome, though their role in MS remains murky (Koch et al., 2013; Küçükali et al., 2014; Zhou et al., 2014).
The field is still trying to parse out which epigenetic changes are associated with MS and what causes them. But researchers see a vast potential to utilize epigenetics for both personalized therapies and preventive treatments (Casaccia-Bonnefil et al., 2008; Koch et al., 2013; Küçükali et al., 2014; Zhou et al., 2014).
Epigenetics 101
Epigenetic changes represent the environment’s effect on the genome. Stimuli such as smoking and aging modulate gene expression without altering the composition of DNA. Epigenetic modulations come in three flavors: DNA methylation, histone modification, and microRNA modification.
- DNA methylation is perhaps the best understood epigenetic modification. It occurs when a DNA methyltransferase (DNMT) enzyme attaches a methyl group to the fifth position of a cytosine nucleotide, silencing the gene. Methylation tends to occur on CpG islands, 300- to 3000-base-pair-long sections of DNA that have more than 50% cytosine and guanine nucleotides. CpG islands are usually located on or around promoter regions, and it’s thought that their methylation prevents transcription factors from binding to the promoter. DNA methylation plays a role in development, cell proliferation, and genomic stability (Casaccia-Bonnefil et al., 2008; Koch et al., 2013; Küçükali et al., 2014; Zhou et al., 2014).
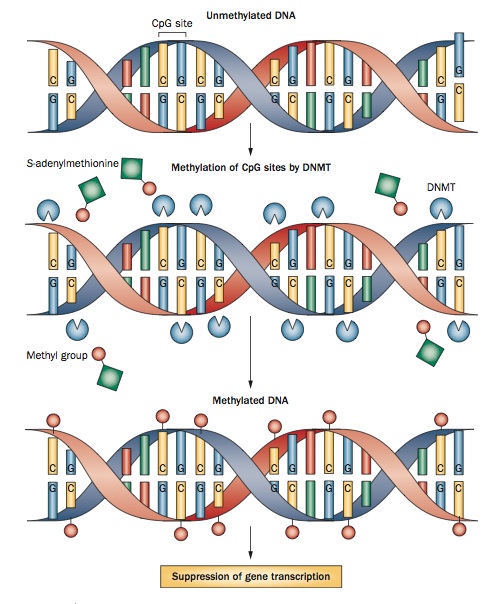
- Histone modification. DNA wraps around histone proteins to package itself into its compact form, chromatin. Histones can be modified in a variety of ways, including acetylation, methylation, phosphorylation, and ubiquitination. These modifications induce changes in the histone that affect the accessibility of the DNA that’s wrapped around it. These changes can either silence or promote the expression of a gene. Among mechanisms of histone modification, researchers have focused most of their attention on acetylation and deacetylation of lysine residues within the histone protein. Generally speaking, acetylation promotes DNA transcription, whereas deacetylation prevents it (Casaccia-Bonnefil et al., 2008; Koch et al., 2013; Küçükali et al., 2014; Zhou et al., 2014).
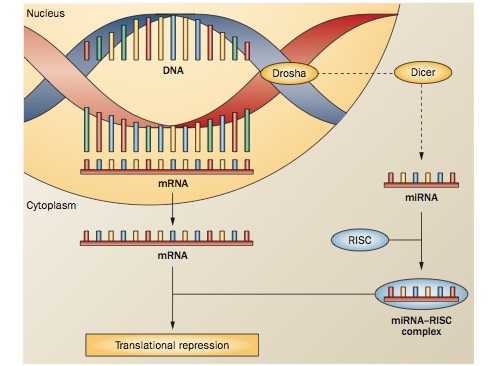
- MicroRNAs (miRNAs) are very small pieces of RNA, roughly 21 to 25 nucleotides long. They can effect change by binding to mRNA and creating an RNA-induced silencing complex. The RISC degrades the mRNA, preventing its translation. miRNAs play a role in many cellular functions, including differentiation and apoptosis (Casaccia-Bonnefil et al., 2008; Koch et al., 2013; Küçükali et al., 2014; Zhou et al., 2014).
Epigenetics in MS
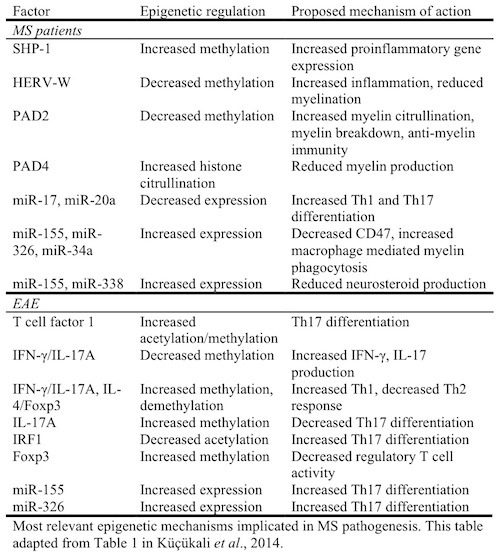
The role of epigenetics in MS is only beginning to come to light. Researchers have identified several mechanisms of epigenetic change in both humans with MS and in the experimental autoimmune encephalomyelitis (EAE) mouse model. The following table summarizes the current research.
“What is it about these people that makes them susceptible and others not? That’s where I think epigenetics comes in,” Rodney Scott, Ph.D., of the University of Newcastle in Callaghan, Australia, told MSDF. “There’s not an overwhelming genetic factor associated with MS. You can identify a genetic factor—it’s located on chromosome 6, the MHC locus—and that clearly does predispose you to MS, but not everyone who has that gets MS.”
Scott’s team has done studies looking at leukocytes in MS patients and healthy controls, and they’ve found notable epigenetic differences between these two groups.
“The epigenetic story is beginning to make a little more sense now, in terms of what are the specific differences. The question now is, what’s causing these differences?” Scott said.
Some of those causes may be easier to identify than others. EBV, for example, is known to stimulate epigenetic changes, and infection with the virus approximately doubles a person’s chance of developing MS. Cancer researchers suspect that EBV leads to hypermethylation of certain promoter regions, resulting in tumorigenesis (Küçükali et al., 2014).
Some preliminary research suggests that in cells infected with EBV, several miRNAs from both the virus and its host are present. In particular, EBV miRNA interacts with miR-142-3p and miR-155. Of those, miR-155 is known to affect T-cell differentiation and promote CNS inflammation in EAE (Küçükali et al., 2014).
Smoking is also known to motivate epigenetic changes in cancer patients through the methylation of tumor-suppressing genes and of brain-derived neurotrophic factor. It also affects miRNA expression and histone changes, though its precise contributions to MS remain unknown (Küçükali et al., 2014).
Of course, not everyone who smokes or contracts EBV winds up with MS. A variety of factors incur epigenetic changes. Perhaps chief among them is aging, according to Patrizia Casaccia, M.D., Ph.D., of Mount Sinai Hospital in New York, NY.
“Age is a major epigenome modifier. We’ve shown this in rats, mice, and humans. Aging does affect the epigenome,” Casaccia told MSDF.
Challenges in epigenetics
There are, of course, many challenges in studying epigenetics. Unlike genome-wide association studies that can be conducted from easily obtained blood or skin samples, epigenome-wide association studies (EWAS) are not as easy. Epigenetic modifications are cell-specific.
“For genetics, if you think of the DNA, it does not matter if you take the DNA from fibroblasts, skin, blood, whatever organ. You’d have the same genetic mutation regardless,” Casaccia said. “But in epigenetics, it’s completely different. A liver cell would be different from a blood cell or a brain cell.”
Additionally, monitoring epigenetic changes in the human brain is also challenging. Samples can only be obtained postmortem, according to Casaccia. The conundrum there is obvious: “How do you know if what you are studying in a postmortem brain is what really happens in life?” she said.
It’s easier to monitor epigenetic changes in immune cells circulating in the blood. But even that is challenging because of the frequency of different cell types.
“There are high frequencies of CD4-positive cells or CD19-positive cells that can be readily isolated from peripheral blood, but rarer forms are too infrequent to readily characterize and study,” Scott told MSDF in an email. “An example of a rare population would be T follicular helper cells that are associated with the production of antibodies, something that underpins autoimmune disease.”
A study of epigenetic modifications in patients with rheumatoid arthritis published in Nature Biotechnology may have a solution (Liu et al., 2013). The team corrected for cellular heterogeneity by applying a series of ad hoc statistical filters to whole-blood samples. In their study they suggest that their method could be applied to all sorts of epidemiological EWAS.
Of course, researchers need a large number of tissue samples to conduct such studies. MSDF’s parent organization, the Accelerated Cure Project, has a repository of biosamples from more than 3000 volunteers. The goal of the ACP repository is to facilitate more studies, such as EWAS, to aid in MS research progress.
The potential for therapy
Because epigenetic changes should be reversible, in theory at least, epigenetics opens an enticing prospect for novel treatments in MS. Enzymes that modulate DNA methylation and histone modifications are easy targets for drug treatment. Currently, though, DNMT inhibitors aren’t specific and can suppress DNA methylation across the entire genome. Since hypermethylation is not considered a widespread epigenetic change associated with MS, it is unlikely that DNMT inhibitors will be useful for MS anyway.
However, inhibiting histone deacetylation shows more promise as an epigenetics-based MS therapy. Casaccia’s team recently published a study in Chemical Biology about a new small molecule, BrDi, that modulates histone acetylation (Gacias et al., 2014). They were able to use the drug to either promote or inhibit oligodendrocyte progenitor cells from maturing into oligodendrocytes.
“This is promising because this is one of the first approaches of addressing with a chemical molecule an epigenome change that occurs with aging.”
Casaccia imagines a world where doctors could use epigenetic knowledge to prevent MS. A doctor could sequence a patient’s genome, understand his or her predisposal to developing MS, and advise diet and lifestyle changes to prevent the epigenetic changes that might trigger the onset of the disease.
“We could give the patient the opportunity to change their lifestyle and give them a chance to avoid the disease,” Casaccia said.
“At the moment, an ‘epigenetic baseline’ must be established,” Jimmy Huynh, Ph.D., of Mount Sinai Hospital told MSDF in an email. “The development of molecules targeting the epigenome holds great hope, as it may be possible to correct epigenetic ‘deviations’ and return a landscape to baseline.”
Raymond Sobel, M.D., of Stanford is less optimistic about the potential for therapy. The prospect of epigenetics-based personalized medicine for MS patients is interesting, but the number of epigenetic changes that any one individual may have must be taken into account.
“Each individual might have factors that are at play, so it would be very difficult to tailor a specific target right into an epigenetic process in people and show that it’s worth doing,” Sobel said to MSDF. The possibility of adverse side effects, especially from using multiple drugs at once, is another factor to consider.
Epigenetics in the future
Before researchers explore therapies in earnest, they still need to lay much of the groundwork toward parsing out which epigenetic modifications are risk factors for the disease and which are results from the disease itself. Eventually, though, epigenetics will aid in diagnosis and prognosis, Huynh said.
Scott believes that technological advances that will reduce the cost and effort of doing epigenetics studies are just around the corner. “I think we’re going to be able to look more closely in the brain where all the action is,” Scott said. “We need to understand what it is that results in the changes associated with MS and the gradual decline in the person’s health over the course of their lifetime.”
Sobel noted that epigenetics might also have the capacity to change MS research itself. The role of epigenetics in MS highlights the disease’s complexity. Since EAE is usually induced in a single event, such as exposure to an antigen, EAE may fail to replicate some of the epigenetic changes involved, potentially limiting its use as an animal model of MS.
Casaccia said she feels that the importance of epigenetics is only just starting to reach the ears of many researchers.
“Ten years ago, there was maybe only my group. Now there are many groups [that study epigenetics in MS]. I think there has been a progressive interest over time. It’s just that sometimes, especially in clinical neurology, concepts tend to trickle down relatively slowly.”
Scott expressed optimism for MS research, saying, “I think we are moving in the right direction. It may not be as quick as people would like, but it’s an awful lot quicker than it has been. There’s great hope in the near- to medium-term future for all MS sufferers.”
Key open questions
- What is the most efficient and effective way to profile epigenome changes in MS?
- What is the viability for personalized epigenetics-based therapies in MS patients?
- How can the MS community better embrace epigenetics?
- Are there any possible workarounds to study epigenetic changes in the brains of living patients?
- How can we better use animal models to study epigenetics?
Disclosures and sources of funding
Rodney Scott and Raymond Sobel report no competing interests. Patrizia Casaccia-Bonnefil receives funding from NIH, FastForward, and EMD-Serono.



Comments
For futher reading please see:
Epigenome-wide differences in pathology-free regions of multiple sclerosis-affected brains.
Huynh JL, Garg P, Thin TH, Yoo S, Dutta R, Trapp BD, Haroutunian V, Zhu J, Donovan MJ, Sharp AJ, Casaccia P
Nat Neurosci. 2014 Jan. Epub 2013 Nov 24. PMID: 24270187.
Epigenetic mechanisms in multiple sclerosis: implications for pathogenesis and treatment.
Huynh JL, Casaccia P.
Lancet Neurol. 2013 Feb. PMID: 23332363