Epigenetic Changes Color the Genomes of People With MS
Brain samples reveal a pattern of DNA modifications that may turn the immune system up and tone repair responses down
The chemical decorations that adorn DNA could sway the onset or severity of multiple sclerosis just as much as the DNA sequence itself, suggests a study published November 24 in Nature Neuroscience. In addition to the genetic sequence that encodes the proteins expressed in each cell, chemical modifications to DNA dictate which genes get turned on and off. The study showed that people with MS display a different set of these modifications—collectively called the epigenome—than do people without the disease (Huynh et al., 2013).
Changes to the epigenome can be sparked by environmental factors such as diet, smoking, stress, and vitamin D levels, so the results of the study support the need for a “more holistic approach” to MS research, said senior author Patrizia Casaccia, M.D., Ph.D., a professor of neuroscience, genetics and genomic sciences, and neurology at the Icahn School of Medicine at Mount Sinai Hospital, New York, NY, in an interview with MSDF. “It’s a very different way to think about the problem,” she said.
Beyond genetics
Genetic mutations have been shown to play some role in MS, as 110 susceptibility loci (at last count) have been identified throughout the genome. Yet the genetic code only tells a small part of the story, Casaccia said. If DNA sequence were the only factor determining susceptibility to MS, “you would expect 100% of identical twins to share the disease,” she said.
Yet only about 30% of identical twins share the disease with affected siblings, compared to the 0.1% chance faced by the general population. People with a parent or nontwin sibling with MS face a 1% to 3% chance of contracting the disorder.
So while the genetic code contributes to MS susceptibility, environmental factors clearly play a strong role, Casaccia said. “And epigenetic modifications are those that allow different patterns of gene expression in response to environmental stimuli,” she added.
The modifications come in many different styles, ranging from direct methylation of DNA to the acetylation of histone proteins that wind around the strands. Diverse as they are, epigenetic changes share a common thread: They alter the expression of nearby genes.
DNA methylation, the most studied form of epigenetic modification, tends to turn down gene expression. Methyltransferases add methyl-group adornments to cytosine residues positioned next to guanine residues. A majority of these so-called CpGs are methylated in the human genome, but clusters of unmethylated CpGs called “CpG islands” exist in the promoter regions of most genes. The nakedness of these islands allows easy access to transcription factors that switch on downstream genes. Wonky methylation patterns have been associated with the development of cancer and other diseases.
A 2010 study looked for epigenetic differences in three sets of twins of whom only one member of each pair had MS, but found no common vein of modifications specific for the disease (Baranzini et al., 2010). For that study, the researchers analyzed DNA from lymphocytes in the blood. Given that epigenetic modifications vary widely between different cell types, Casaccia’s team decided to look for an epigenetic signature of MS in the tissue most vulnerable to the disease: myelinated brain tissue.
An epigenetic signature
The researchers collected postmortem brain tissue from 28 multiple sclerosis patients and 19 controls. The samples, which were obtained from the UK Multiple Sclerosis Tissue Bank and the Human Brain and Spinal Fluid Resource Center at UCLA, were selected only after stringent screening for high-quality genetic material and a short time since death (31 hours or less). The researchers isolated DNA from the normal appearing white matter (NAWM)—myelinated brain tissue that appeared free from the pathology of MS disease—in an attempt to avoid confusing epigenetic changes caused by the pathology of the disease with those that may have brought it on in the first place.
The team isolated DNA from the brain samples and analyzed genome-wide methylation patterns using a specialized microarray chip. After correcting for the effects of age and gender, the team identified 220 hypomethylated regions and 319 hypermethylated regions in the genomes of people with MS as compared to normal controls. Many of the regions fell within previously identified gene enhancers: segments of DNA known to control levels of gene expression.
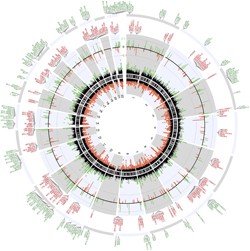 A closer look at the genes associated with the differentially methylated regions (click for larger image) revealed an intriguing pattern. Hypomethylated regions were detected around several immune response genes, while genes associated with oligodendrocyte function were among those containing hypermethylated regions.
A closer look at the genes associated with the differentially methylated regions (click for larger image) revealed an intriguing pattern. Hypomethylated regions were detected around several immune response genes, while genes associated with oligodendrocyte function were among those containing hypermethylated regions.
To confirm that the differences in methylation between the two groups had real consequences for gene expression, the team performed a genome-wide transcriptional analysis using RNA isolated from the brain samples. When they compared the methylation data with the transcript data, the investigators found that genes containing hypermethylated regions tended to be turned down (P = 9.8 × 10-5), whereas those containing hypomethylated regions tended to have increased expression (P = 1 × 10-18).
The researchers confirmed the findings in a second cohort of brain samples collected from the Lerner Research Institute at the Cleveland Clinic in Ohio, which showed similar patterns of methylation and expression differences between the brains of people with MS and those of controls. The team even correlated the methylation changes with protein expression in a subset of the genes.
While the changes in methylation were subtle in many cases, the effects were widespread and clearly affected gene expression, Casaccia said. “Little changes in many regions become cumulative,” she said. “If you have all of these changes in a patient that already has genetic susceptibility, then there could be a tendency for the disease to take over.”
Despite the thinly painted epigenetic changes the researchers reported across the genomes of people with MS, other scientists who were not involved in the study expressed confidence in the results. “They made an incredible effort to use every tool available to ensure that their identification of small changes was valid,” said Gwenn Garden, M.D., Ph.D., a professor of neurology at the University of Washington, Seattle, in an interview with MSDF.
Garden, who studies factors (including epigenetics) that may switch on inflammation in the brain, said the study’s results may prove useful in identifying targets for treatments, but that much work will be needed before researchers understand which environmental factors lead to the epigenetic changes.
“This work is really an interesting start,” said George Ebers, M.D., a professor of neurology at the University of Oxford, in an interview with MSDF. Yet Ebers added that the analysis does not answer the question of whether the epigenetic changes triggered disease. “It’s always a question of what the changes actually mean,” Ebers said. “Are they cause or effect?”
Casaccia acknowledges that the study’s results don’t reveal which environmental factors trigger the methylation changes or how those changes may lead to disease—a complex thicket of questions she hopes to tackle along with other scientists in the MS research community.
“We need studies that will explain the amount of influence that diet, smoking, and these other environmental changes actually have on the disease,” she said. “Ideally, you want one drug that does it all: affects the immune system and causes repair. But maybe it’s not reasonable, and we really need to understand the factors.”
Key open questions
- Do epigenetic changes trigger MS?
- What environmental factors trigger the epigenetic changes associated with MS?
- How does genetic susceptibility influence epigenetic susceptibility to MS and vice versa?
- Why are certain genes targeted for epigenetic modification?
- If MS is an epigenetic disease, how will this change approaches to treatment?
Disclosures
Seed funds from the Icahn School of Medicine and grants from the National Institutes of Health supported this study. In addition to an NIH Fellowship, first author Jimmy L. Huynh is a recipient of a scholarship from the Foundation of the Consortium of Multiple Sclerosis Centers' MS Workforce of the Future. Drs. Casaccia, Garden, and Ebers reported having no financial disclosures.


