Probing T Cell Gene Regulatory Networks for Early Signs of Disease
A network model of gene expression during human T cell differentiation, combined with results of genome-wide association studies, helps identify potential disease markers in asymptomatic patients with MS
In relapsing-remitting multiple sclerosis (RRMS), normally protective immune T cells transform into effector cells that attack the myelin in the brain and spinal cord. Because the mobilization and maturation of T cells precedes symptoms in RRMS, understanding the initial events in immune activation could provide biomarkers for presymptomatic diagnosis.
In a paper published November 11 in Science Translational Medicine (Gustafsson et al., 2015), Mikael Benson, MD, of Linköping University in Sweden and colleagues outline a strategy to identify early regulators of T-cell maturation. Their work pinpointed three key transcription factors involved in T-cell differentiation and enriched in disease-associated polymorphisms, and whose activity is changed in a number of immune-related diseases, including multiple sclerosis.
The transcription factors—GATA3, MAF, and MYB—were also altered in asymptomatic MS patients in remission, which the researchers used as a proxy for early disease.
While it’s not yet clear how the activity of biomarkers during apparent MS remission will ultimately relate to presymptomatic diagnosis or prognosis, the study provides a general scheme for identifying early T-cell events that may be applicable to many diseases.
Previous studies from the Swedish group and many others have employed microarray technology and transcriptome profiling to identify alterations in gene expression programs in T cells from different diseases, including MS. But those studies tackled established disease and so could not identify the key events leading to those changes.
To identify important initial steps, associate professor Mika Gustafsson, PhD, doctoral student Danuta Gawel, and colleagues first created a comprehensive catalog of gene expression changes during differentiation of normal human CD4+ T cells. Those changes give rise to helper (TH) and regulatory (Treg) cells implicated in autoimmune diseases, including MS. Starting with naïve CD4+ T cells from healthy donors, the researchers performed whole transcriptome sequencing repeatedly as the cells matured into one of four major T cell subtypes—TH1, TH2, TH17, and Treg—in culture.
The serial expression data revealed which transcription factors were differentially expressed during T cell maturation. To see which might be related to disease, the researchers looked to data from human genome-wide association studies. They asked which factors harbored disease-related variants in studies of multiple autoimmune disorders. Disease-causing variants most often affected TH1/TH2 maturation, they observed.
Focusing on that pathway, the investigators constructed a mathematical gene regulatory network model of early TH1/TH2 differentiation. The model accounted for 6112 interactions of 378 transcription factors and 1563 mRNAs. The investigators extensively validated the model with analysis of transcription factor binding, transcription factor knockdown experiments, and additional expression profiling.
From that complex web of data, the researchers prioritized for further study transcription factors that appeared to be network hubs—factors that affected expression of many genes—and that harbored disease-associated genetic variation. This led to three transcription factors, namely GATA3, MAF, and MYB.
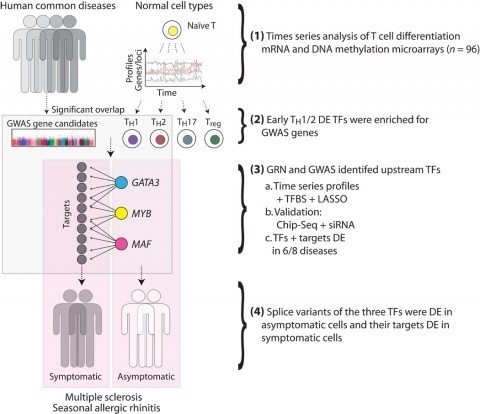
Using existing expression-profiling data from multiple diseases, Benson and co-workers found evidence that the activity of all three transcription factors was altered in T cells from patients with symptomatic MS and seven other diseases including seasonal allergic rhinitis (SAR), rheumatoid arthritis, lupus, and others.
To model early MS, the researchers analyzed T cells from 14 asymptomatic RRMS patients who had been relapse-free for at least 3 months (patients in remission). The patients had been diagnosed with RRMS according to the McDonald criteria and had a median Expanded Disability Status Scale score of 3 (range 0-4) and a median MS Severity Score of 2.7 (range 0.21 to 4.95).
Compared to healthy controls or relapsed patients, the people in remission had clear changes in expression of GATA3, MAF, and MYB. Symptomatic patients showed differential expression of GATA3, MAF, and MYB target genes. Differences in factors or their target genes were also found in people with SAR who were asymptomatic, symptomatic, or successfully treated.
 “We used the symptom-free stages in MS as a model of early disease, but obviously you will need to study large numbers of healthy individuals for long periods of time to study if these markers are useful to detect disease before symptoms,” says Benson, a professor of pediatrics and systems medicine.
“We used the symptom-free stages in MS as a model of early disease, but obviously you will need to study large numbers of healthy individuals for long periods of time to study if these markers are useful to detect disease before symptoms,” says Benson, a professor of pediatrics and systems medicine.
"They are tackling important questions,” says Alexander Marson of the University of California, San Francisco, who was not involved in the study. “Are different genetic programs involved in different disease stages, and can we find biomarkers of disease in people who are asymptomatic?"
The study is commendable for its ambitious scale and for generating data that will be useful going forward, Marson says. “It’s producing a lot of information on gene expression programs in human T cells, and they’re finding overlaps with gene signatures that have come out of the genome-wide association studies for immune-mediated diseases. The more data that we have in the field, we’ll start to really be able to hone in on connections of how T cells are working in healthy states, and how they go awry at various stages of disease progression,” he told MSDF.
For Benson, a larger problem than early detection for MS is the individual variability in response to treatments. Using the same methods, his group has already identified a set of T cell biomarkers that predict nonresponse to natalizumab treatment in MS patients, which he believes could be used in the clinic in the near future (Gustafsson et al., 2014).
Key open questions
- What is the role of the three transcription factors’ (GATA3, MAF, and MYB) gene expression programs in T cells in the actual initiation of MS, as compared to remission?
- How might a deeper understanding of gene regulatory networks behind T helper cell differentiation shed light on the pathophysiology of MS?
Disclosures and sources of funding
Gustafsson et al.: This work was supported by the Cancer Fund, the Swedish Medical Research Council, the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research 2012–2017, the Sigrid Jusélius Foundation, the Generalitat de Catalunya AGAUR, the Spanish Association Against Cancer, and the Spanish Ministry of Health. The authors declare that they have no competing interests.
Marson’s lab receives some research support from Epinomics and has a provisional patient on some CRISPR/Cas9 genome editing technology.
Edited by Linda Felaco.


