Gene Variant Associated with Non-Response to Interferon β
A variant of a gene called SLC9A9 appears significantly more often in people with MS who fail to respond to interferon β therapy
In a meta-analysis involving three independent cohorts of patients, researchers have identified a single-nucleotide polymorphism (SNP) strongly associated with nonresponse to therapy with interferon β (IFNβ). While other genome-wide association studies have identified more than 150 SNPs associated with susceptibility to multiple sclerosis (MS), this appears to be the first to identify a genetic association to therapy response.
Published online ahead of print by Annals of Neurology on April 25, 2015 (Esposito et al., 2015), the study was led by Federica Esposito, M.D., Ph.D., of the San Raffaele Scientific Institute in Milan, Italy, working in the lab of Filippo Martinelli Boneschi, M.D., Ph.D. The Milan group collaborated with a group from Boston led by Philip L. De Jager, M.D., Ph.D., from the Ann Romney Center for Neurologic Diseases at Brigham and Women’s Hospital.
The team first identified the SNP in a cohort of 146 patients from Milan, all of whom had relapsing-remitting MS and all of whom were taking one of the three forms of IFNβ (brand names Avonex, Betaferon, and Rebif). Of those patients, 74 responded to treatment, 42 failed to respond, and the remaining 30 were classified as partial responders.
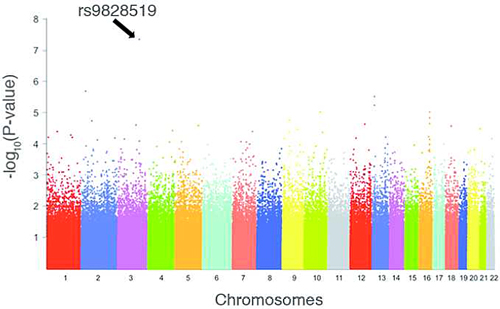
After evaluating more than half a million autosomal SNPs, only a single one, called rs9828519G, exceeded the previously defined threshold of statistical significance: a P value < 5 × 10-8. In this initial cohort that allele was strongly associated with nonresponse to IFNβ therapy, with the astounding odds ratio of 5.46 (95% CI = 2.89-10.33).
In an interview with MSDF, De Jager put this odds ratio into context. “We now have a pretty good idea of the genetic architecture of human traits, whether it’s related to MS or more broadly to all sorts of human traits,” he said. “In general these variants have very modest effects on these traits, with odds ratios between 1.05 and 1.2 or maybe 1.3.”
The team attempted to replicate those results in two additional cohorts, one involving 281 MS patients enrolled at Brigham and Women’s Hospital in Boston, and the other involving 325 patients from seven MS centers in France.
In a well-known game-theory effect called the winner’s curse, the associations between rs9828519G and IFNβ were considerably more modest in the replications than in the initial cohort. The odds ratio from the U.S. cohort was 1.97, for example, and that from the French cohort was 1.33. In the combined meta-analysis of the three cohorts, the overall odds ratio was less astounding than the initial cohort but still quite impressive at 1.48 (95% CI = 1.18-1.86) with a P value of 7.78 × 10-4.
The gene
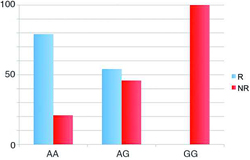 The SNP rs9828519 appears to be associated with a gene called SLC9A9. Among the original cohort, not a single patient homozygous for the G allele of rs9828519 responded to IFNβ. Among those who were homozygous for the A allele, 20.9% were nonresponders. And among those who were heterozygous, with one copy each of the A and G alleles, 45.9% were nonresponders.
The SNP rs9828519 appears to be associated with a gene called SLC9A9. Among the original cohort, not a single patient homozygous for the G allele of rs9828519 responded to IFNβ. Among those who were homozygous for the A allele, 20.9% were nonresponders. And among those who were heterozygous, with one copy each of the A and G alleles, 45.9% were nonresponders.
SLC9A9 seems to be expressed in a number of tissues, including the T helper cells Th1 and Th17, which are well known to be involved in inflammation in MS.
“Not a whole lot is known about [SLC9A9],” De Jager told MSDF. “By looking at its sequence it’s a sodium/hydrogen exchanger. So basically it’s a channel protein and it helps to regulate the pH of some of the intracellular vesicles that are important in the final modification of proteins prior to [transport] to the cell surface. … But exactly how this gene is involved in these processes is not yet clear. So that’s where much of the biology remains to be done, to see exactly why changing the pH of these compartments would affect response.”
This gene family has been implicated in other neurological disorders. “There are actually a couple of groups who are studying this gene and a related gene called SLC9A6 in autism,” De Jager said. “We’re actually talking to these colleagues about what they have found so far and seeing whether there is some connection between autism and MS in terms of the mechanism of these genes. But at this point it’s too early to say whether there’s any sort of connection or [whether] it’s just chance.”
Implications
Asked for independent comment on the new study, Pierre-Antoine Gourraud, Ph.D., of the University of California, San Francisco, told MSDF, “It’s probably one of the first very credible pharmacogenetic studies in MS that’s really found something. It’s fairly important.”
Gourraud pointed out that studies intended to find genetic associations with disease susceptibility present fewer challenges than studies hoping to find genetic associations with treatment response. “In genetic studies the job was probably easier because we just had to agree on who has MS and who [does not have] MS. In here we have to agree on what are the patients who are responding over time and who are not responding over time. And that’s way more difficult to define, and I think it really calls for more worldwide collaboration to define, to have standards in the way we assess progression and phenotype in multiple sclerosis.”
In classifying treatment response, the investigators defined responders as those patients who had no relapses and no new T2 lesions or gadolinium-enhancing lesions within 2 years of follow-up. They defined nonresponders as: (1) those patients whose annualized relapse rate decreased less than 50% compared to the 2 years before treatment, or (2) by the appearance of a certain number of new T2 or Gd-enhancing lesions within 1 or 2 years. And they defined partial responders as those patients who fulfilled neither the definition of responder nor the definition of nonresponder.
While praising the study, Gourraud also expressed some reservations. “I would have hoped for something a little more robust in terms of replication of the effect,” he said.
Gourraud added that the discovery of the gene variant doesn’t reveal much about MS pathogenesis. “It’s a very common variant, so it doesn’t tell you much about really a major effect that would be driving the disease or the response to the disease,” he said. “It shows that it’s a very complex disease, not only in the susceptibility to MS, in the heterogeneity of multiple sclerosis progression, but also in the response to treatment.”
De Jager pointed out that the discovery of the gene variant will have no immediate effect on treatment choices. “By itself this variant is not sufficient to identify people who will have a relapse or those who won’t,” De Jager said. “So to be clear there’s no clinical utility to measuring the variant at the current time. That said, it is a piece of information that is robust. It needs to be validated further, but assuming that it’s validated, we can then [combine other pieces of information] to measure aggregate estimates of response.”
Gourraud agreed. “I don’t think it will be of extremely clear usefulness in the clinic, yet it’s very interesting to better understand who are the responders to the treatment. Maybe the effect will be part of a compound criterion to decide if somebody is responding or to decide about the propensity to be a responder.”
Key open questions
- What is the role of SLC9A9 in MS pathogenesis?
- What other genetic associations to treatment response will be discovered using this approach?
- How can this information be used to tailor MS treatments to individual patients?
Disclosures and sources of funding
Study authors disclosed potential conflicts of interest with a variety of pharmaceutical companies including Almirall, Bayer-Schering, Biogen, Biogen-Dompè, Chugai, EMD Serono, Excemed, Genzyme, GlaxoSmithKline, Nasvax, Novartis, Sanofi-Aventis, Teva Neuroscience, and Vertex. Pierre-Antoine Gourraud is a member of the MSDF Scientific Advisory Board. Philip De Jager is a member of the Scientific Advisory Board of the Accelerated Cure Project, MSDF’s nonprofit publisher.


